DEE2 2025 updates: growth and development
DEE2 is a database service I co-founded in 2015 aiming to provide uniformly processed gene expression profiles for each and every RNA-seq dataset in NCBI's Sequence Read Archive. After some major revisions, we published the database/service journal article in 2019. Over time, DEE2 has grown dramatically, with the number of datasets and total number of counted reads, as seen in the tables below.
Here, I'll walk you through the growth of the database over time and the new features we've added.
Growth of DEE2
DEE2 continues to ingest new metadata from NCBI SRA and GEO, and the growth of this metadata set has caused us a lot of issues over time. In the early years of DEE2 we used SRAdb, and then it became too large for its design, so we sought different solutions. Currently we are using pySRA and only fetching quarterly. The size of each request has been a challenge, with human and mouse requests of annual dumps exceeding the 64 GB RAM of our backend workstation! So that was broken down to 365 separate requests per species. It is a bit slow, but much more reliable. Anyway, the number of datasets, roughly equivalent to samples has exceeded 1.3 million in our latest update (Jan 2025; Table 1).
Table 1. The number of datasets
The number of sequence tags is also increasing gradually, In 2025 we reached 16.5 trillion data points (Table 2). This makes DEE2 larger than ARCHS4 and Recount3, although these three datasets have a different design and priority.
Table 2. Total number of DEE2 sequence reads.
These data are available for bulk download here. For SRA studies with more than four samples, we have provided those data as bundles, which is a convenient way of accessing the data if you already know the SRA project accession number (usually starts with SRPxxx; See Table 3). Of these 44k bundles, 3.8k were added in the past two weeks.
Table 3. Number of project bundles available.
The backlog
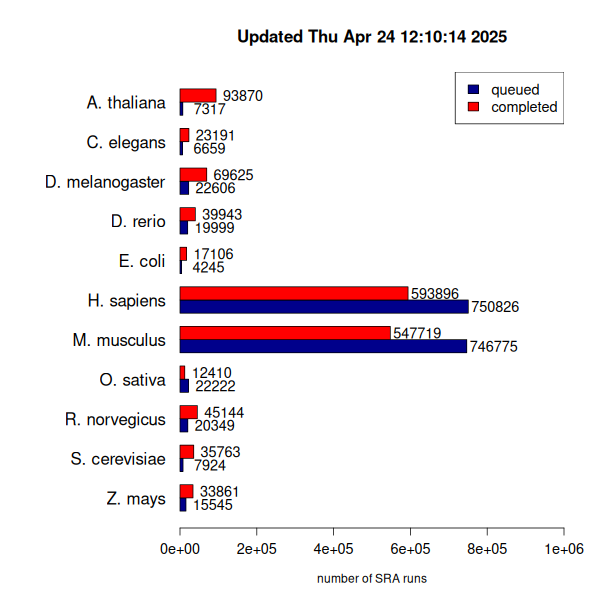
While DEE2 is growing gradually, we have tried to maintain a high fraction of completed jobs. This has been easier for organisms like C. elegans and D. melanogaster where the small genome size makes processing less computationally intensive and the datasets are generally smaller. These organisms also have fewer submissions to SRA as compared to human and mouse. The backlogs of human and mouse datasets (See Figure 1) is something that is difficult for us to stay on top of with the current availability of compute resources, and even if we were able to process these 1.5 M datasets in the near term, we would require upgraded data storage, as our current 4 TB webserver and 8TB workstation computer are at 82% capacity. This would require significant financial resources or a better approach to compression and data retrieval - we are looking at whether hdf5 data structures can work in our case.
Figure 1. Number of available datasets in DEE2 and the size of the job queue.
New species
The biggest update to be announced here is the inclusion of two new species rice (Oryza sativa) and corn (Zea mays). These join Arabidopsis as the only plant species in the database, and we have made available 6k rice and 33k corn datasets already. This was only possible with major support from Dr Wen-Dar Lin's team at Institute of Plant and Microbial Biology who processed all of these datasets, and we are very grateful for their massive contribution. The rationale behind including these is that they had a high number of RNA-seq datasets in SRA and that our database was underrepresented in plant species, in particular monocotyledonous plants which are a major source of food, fuel and fibre. Other organisms that are on our candidate list include Plasmodium falciparum, a species of malaria and Macaca mulatta, rhesus macaque, which is used widely in research due to similarity with human. Macaca mulatta has 49,592 datasets advertised on SRA while Plasmodium falciparum has just 9462.
Changes to the processing pipeline
While we can't make major changes to the pipeline that alter the data output, we have made some quality of life improvements to make things run smoother and more intuitive.
Improved syntax
The first one is the change to the syntax for running the pipeline. Instead of having a mix of positional and declarative options, they are all declarative. See the new help page:
$ docker run -it mziemann/tallyup:dev
tallyup is a dockerised pipeline for processing transcriptomic data from NCBI SRA to be included in the DEE2 database (dee2.io).
Usage: docker run mziemann/tallyup <-s SPECIES> [-a SRA ACCESSION] [-h] [-t THREADS] [-d] [-f FASTQ_READ1 FASTQ_READ2] [-v]
-s Species, supported ones include 'athaliana', 'celegans', 'dmelanogaster', 'drerio', 'ecoli', 'hsapiens', 'mmusculus', 'osativa', 'rnorvegicus', 'scerevisiae' and 'zmays'
-a SRA run accession, a text string matching an SRA run accession. eg: SRR10861665 or ERR3281011
-h Help. Display this message and quit.
-t Number of parallel threads. Default is 8.
-d Sequence data is downloaded separately in SRA archives (this is the most efficient way). This option will process all SRA archives with .sra suffices in the current working directory.
-f1 User provided FASTQ files for read 1.
-f2 User provided FASTQ files for read 2.
-v Increase verbosity.
EXAMPLES
Provide SRA run accessions run on 6 threads:
docker run -it mziemann/tallyup -s ecoli -a SRR27386505,SRR27386506 -t 6
Process SRA archives with verbose logging:
prefetch SRR27386504
docker run -it -v /dee2:/dee2/mnt mziemann/tallyup -s ecoli -d -t 8 -v
Process own fastq files with 12 threads:
docker run -it -v /dee2:/dee2/mnt mziemann/tallyup -s ecoli -f1 ecoli_SRR27386502_1.fastq.gz -f2 ecoli_SRR27386502_2.fastq.gz
Return data to host machine:
docker cp `docker ps -alq`:/dee2/data/ .
OUTPUT
If SRA archive files are provided, then the pipeline will create a zip file in the host working directory that contains the processed RNA-seq data.
If accession numbers or fastq files are provided, then the command is required to retrieve the processed RNA-seq data.
Better control of parallel threads
Secondly, the number of parallel threads is better controlled. The -t option specifies the number of parallel threads rather than using system `proc`, which is problematic for a number of reasons. For example on shared HPC system with 48 threads the pipeline would assume all threads are available, but in reality it is much less (a typical job might have only 12-16 threads). The pipeline has some single threaded bottlenecks, so keeping the number of parallel threads to 8-16 is the best range. This means that a single Xeon or EPYC/Threadripper workstation can run 3-6 ongoing parallel worker jobs of 8-16 threads each, which is very efficient for data processing. I ran a test of this new parallelization on a 16 Core AMD Ryzen Threadripper PRO 5955WX system with two datasets (see Table 4 and Figure 2).
- D. melanogaster, SRR25512078, 1.4 G, 26.7M reads
- H. sapiens, SRR3647493, 3.6 G, 25.8M reads
Table 4. Testing how adding more threads speeds up data processing. Values show time in seconds.
Figure 2. Execution time for two datasets with 4 to 32 parallel threads.
Adding more parallel threads doesn't really work beyond 12 threads. So keep DEE2 processing jobs to 12 or less. If you have more available RAM you can run them at 8 threads each.
Official support for Apptainer
Docker has been great for us, especially on workstations where we have admin control, but for shared systems like HPC that forbid docker, then we have had limited options. I originally tried singularity in 2020, but it was problematic and we moved to udocker. That worked well for me for a long time but would lead to the home directory getting filled up quickly and HPC admins getting irate at the rate of data storage being used. So based on advice from our HPC admins at our institute, I used Apptainer. My first attempt, using the SIF file as the image wasn't a success as that lead to a major overhead in data I/O which made the processing extremely slow. I have had better luck when the image is a simple folder structure. If you are interested in processing some datasets, then I have made a walk-through guide. It will get you started very quickly and with only a few commands.
Final thoughts
- HDF5 for efficient storage and retrieval
- Some new species, like rhesus monkey and malaria
- A new dedicated server at our institute with Threadripper 5995WX 64 cores, 128 threads
Maintaining DEE2 has been a major challenge and a truly huge amount of work, but it is also rewarding to see that it is helping people access these valuable datasets and answering important biological questions. I love seeing what creative applications you have for this dataset. Thank you for your support of the project!