DEE2 project update October 2020
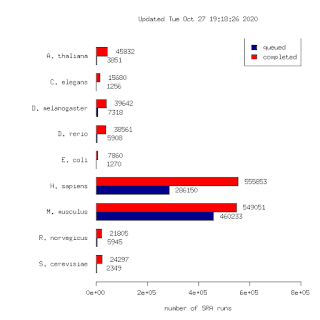
Bit of a thread for some updates to the DEE2 data set. It's a resource of uniformly processed RNA-seq data free to use under a permissive GPL3 licence. Find it at http://dee2.io Yesterday a batch of 117k human runs were uploaded. This brings the total number of runs to 1,298,581. To my knowledge this is the largest such data set in the world. This is 10x larger than our first release in 2015! (125k runs) The number of SRA projects with completed data analysis bundles is 32692 Accesible here: http://dee2.io/huge/ The getDEE2 package is the recommended way to access this data if you are familiar with R. You can access individual runs or enrire data bundles with the various functions. The pkg is part of the latest BioC release out today https://www.bioconductor.org/packages/devel/bioc/html/getDEE2.html The button for redirecting DEE2 data directly to Degust is broken and we are looking for a fix. For now you will need to download the data to the disk and upload to the Degu...

