Is paired end RNA-seq better than single-end for gene-wise gene expression analysis?
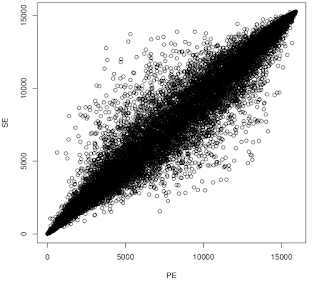
Something I've wondered about is whether for RNA-seq it's worth forking out the extra cost of sequencing both ends as opposed to single end. To test this, I went back to a paired end data set present in GEO ( GSE55123 , 2x 36bp), cleaned the data with Skewer, then mapped the reads with STAR in either paired-end mode or single-end mode (using just read 1). I then used featureCounts to quantify number of tags aligned to each gene. I excluded genes with fewer than 10 reads per sample on average. Then I ran edgeR at Degust to identify differentially expressed genes (DEGs@FDR<0.05). I used a shell script to quantify the overlap in DEGs. Then I ranked them based on the p-value from most up-regulated to most down-regulated and compared their positions in the rank. Here's the result of the overlap analysis. You can see that PE fastq detected more genes but identified fewer DGEs than SE. Detected in PE:15919 Detected in SE: 15275 Detected in both:14750 Detected in eit...