Gene name errors: Redux
Our latest article "Gene name errors: Lessons not learned" previously @biorxiv_genomic has just been published in PLoS Computational Biology (link here). In this post I'll walk you through why it is so important to how computational biology is done. If you are a regular GenomeSpot reader you are probably well aware about how Excel mangles gene names. So why the need for a 2021 update?
Well we thought that the broader genomics community would know about it by now. It has been nearly 20 years since Zeeberg et al's paper on the issue, and five years since our article in Genome Biology which got picked up in a few media outlets and led to SEPT, MARCH and DEC genes being renamed.
So with Mandhri Abeysooriya an outstanding Masters student at Deakin University, we set out to see whether gene name errors in supplementary data files was still an issue. We also had massive assistance from Megan Soria and Mary Kasu.
When designing this new work, we wanted to make it bigger and better than the 2016 study, so we decided to analyse ALL genomics related articles we could get our hands on from NCBI Pubmed Central. We decided to screen articles in the period 2014 to 2020 because that would highlight whether researcher behaviour and excel use mighthave changed in the period after 2016
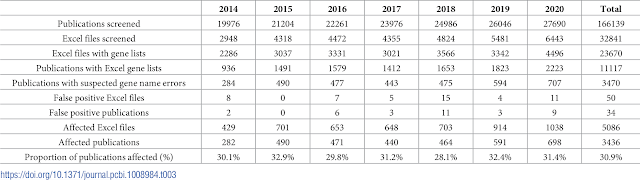
All up we looked for supplementary Excel files in 166,139 PMC articles and 11,117 of these had Excel files with gene lists we could examine. The script we used to perform the error detection was slightly improved so that it also picked up 5-digit numbers (internal date format) as errors, something we overlooked previously. The new code is uploaded to GitHub here.
Of this set of 11117 articles, 3436 were affected by gene name errors which was actually higher than we previously estimated, at approximately 31% of articles with excel gene lists.
This figure is quite a bit higher than the 2016 article and not what we were expecting at all. We attribute this discrepancy to the inclusion of internal date format errors as well as the differnt sampling approach used. Instead of just 18 major journals last time, here we have articles from 741 journals.
The team opened up each of the spreadsheets to confirm the errors, which took a team of 4 people a few weeks to do. There were a few false positives, but overall the script was working as expected.
What shocked us was that when looking at how frequently these errors occurred, the high impact journals like Nature, Cell and PNAS were at the top of the list! Clearly these journals didn't get the 2016 memo!
While we were opening all these spreadsheets, we found a number of edge cases where excel was doing really weird things to some gene names like AGO2, MEI1 and TAMM41. These turned out to be related to the locale settings of the software, recognising these as dates!
There were other cases where protein names like jun-1 were converted to May 31st, so we're presuming that Excel is interpreting jun-1 as the month of June minus 1. Truly bizzare.
What these edge cases demonstrate is that simply changing SEPT, MARCH and DEC gene names isn't going to solve the issue because there are problematic gene names in other locates. Also those protein names like jun-1 will need to be fixed too.
Also, this isn't a problem limited to human and mouse. All eukaryotic kingdoms represented in Ensembl has excel genes, which you can explore in the supplement.
Yes it is ironic that our article has XLS supplementary files, but understand this is the best was to demonstrate how the errors look. We still recommend researchers use TXT, CSV and TSV formats to share their genomics data. We also share a set of recommendations that researchers can use to avoid gene name errors. (Thanks to the reviewers who recommended this)
From the evidence we see, it is likely that gene name errors will continue frustrating bioinformaticians well in to the future. Excel is just so engrained in the way science is taught and done that it will take generational change for it to be eradicated. I joke that Excel will keep me employed forever as there's always new error modes being discovered. Just in the few hours after tweeting this Larry Parnell @larry_parnell replied that he saw the Apple spreadsheet "Numbers" converting gene names into currencies! Lord give me strength.
Until journals start mandating that analytical code be provided for analysis of high throughput data, it's unlikely that Excel will be unseated as the analytical workhorse it is now.
That's not to say that workflows like Rmarkdown and Jupyter notebooks are perfect; they're not. But at least shared code can be reproduced, and more readily audited.
Before I go I need to thank Neil Saunders @neilfws - his blog post was the motivation behind the 2016 study and the title of this update. **THANKS!!**
I would also like to thank the reviewers they really did provide criticism that was constructive and helped the quality of the end product.
I also set up a bot which scrapes PMC each month for errors, so you can see which journals are the worst offenders, see the reports here.