Update on DEE2 project for Jan 2018
Today I'd like to share some updates on the DEE2 project, which I wrote about in an earlier post. The project code can be viewed on github here.
Pipeline and images
As the pipeline was recently finalised, I was able to roll out the working docker image. To facilitate users without root access, this image was ported to singularity. This took a lot of effort and some expertise from our local HPC team to get things working (many thanks to the Massive/M3 team). The singularity image is available from the webserver (link) and instructions for running it are available on github here. I have started testing a heavyweight singularity image, which includes the genome indexes, which will be more efficient for running jobs with large genomes and will make it available once testing is complete.
Queue management
It may sound simple to write a script to determine which datasets have been completed and add new datasets to the queue but when taking about tens of thousands of datasets it's quite a challenge. Several updates have recently gone into the R script that checks incoming data and manages the queue. Initially I wanted this script to also perform matrix construction, but I am finding R to be too slow an memory hungry for this purpose so I'm reverting to python or shell tools to do that. The queue script will be run every 6 to 12 hours, updating the queue on the webserver. Queue management on the webserver is also updated, so that the number of times a certain dataset is allocated is recorded, so that allocations are spread more evenly. This should result in more efficient allocations.
Data throughput
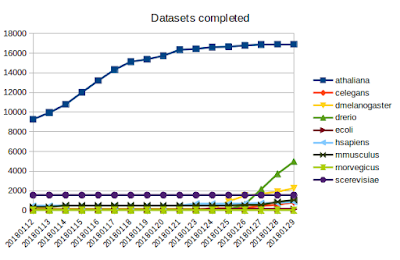
Since the start of the year, I've focused compute power on getting 95% of the Arabidopsis datasets completed which I think we've now achieved. These datasets were processed by our Nectar cloud servers as well as a home server. On the 26th Jan, we got the singularity image working on the M3 cluster, focusing on D. rerio with 4-6 individual servers, as you can see in the graph below, it is progressing rapidly.
But we are FAR from finished, as you can see from the size of the queue below, mouse and human datasets dwarf the others. Which is why we're investigating all avenues to add processing power. This will include spot instances on AWS and preemptive Google cloud, and there will be step-by-step guides for new users who want to use cloud computing to process their RNA-seq data. We are also keen to gain an allocation at the NCI supercomputer.
Data provision and webserver
I mentioned that the way the bulk matrices are generated is changing, indeed I found this to be a bottleneck and unfeasible for more than a few thousand datasets. Using the current R script, matrix generation of the Arabidopsis data crashed the 128GB server in multi-core mode but took >3 days in single core mode so for the moment it will be done using unix tools like split and paste (about 7 minutes with my prototype script). Matrix generation will be run daily but might have to be less frequent as more data is processed. The current matrices can be found at the bulk data page here.
Currently the search function is not operation so the data is only available from the bulk data page. I will be working on developing the user interface over the next 2 months, which I think will resemble DEE v1 but with some modifications. Firstly I will have another crack at implementing a search engine to identify datasets of interest from the metadata. And secondly I will be making cosmetic changes to the site to make it more attractive. Currently the webserver is (here) but will be mapped to a new domain once the data processing is near completion.
How you can use DEE2 in your work
There are a number of ways you can use DEE2 already. You can download and play with the bulk data matrices located here. If SRA/GEO datasets of interest are missing - as is likely the case at this early stage in the data processing phase, then you can use the docker image on your server or spin up a cloud server and follow the steps outlined in the README.md. One of the nicest features of the image is that it can process your own fastq files. This is perfect when comparing your own RNA-seq data in fastq format to some public data. Finally the singularity image will be of use for users without sudo or root access, for instance HPC users.
We fully welcome contributions to compute time so if you have an idle machine consider leaving it to process data using this command, replacing <species> with your favourite model organism:
or
nohup bash -c 'while true ; do docker run mziemann/tallyup <species> & tailf nohup.out ; done'
Also contributions to code base are welcome as are your suggestions and comments below.