Update on DEE2 project for Jan 2018
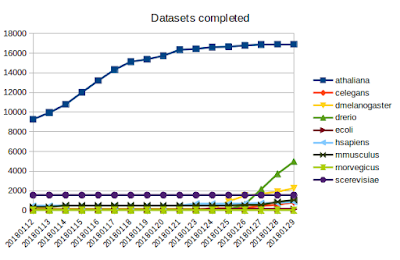
Today I'd like to share some updates on the DEE2 project, which I wrote about in an earlier post . The project code can be viewed on github here . Pipeline and images As the pipeline was recently finalised, I was able to roll out the working docker image. To facilitate users without root access, this image was ported to singularity. This took a lot of effort and some expertise from our local HPC team to get things working (many thanks to the Massive/M3 team). The singularity image is available from the webserver ( link ) and instructions for running it are available on github here . I have started testing a heavyweight singularity image, which includes the genome indexes, which will be more efficient for running jobs with large genomes and will make it available once testing is complete. Queue management It may sound simple to write a script to determine which datasets have been completed and add new datasets to the queue but when taking about tens of thousands of ...