Pathway analysis: DAVID versus GSEA
Pathway analysis has become such a common procedure in bioinformatics, especially in studying gene expression. If you look at a survey of recent papers it seems that there is a bunch of ways that it can be done. In this post, I'll discuss the differences between two commonly used tools; DAVID and GSEA.
The concepts behind the two algorithms are very different. DAVID determines overlaps between user-supplied gene lists and the curated databases, looking for overlaps that are bigger than that expected by random chance. You can improve the accuracy of the algorithm by providing a background file that contains all genes that were considered/detected in the experiment. The user-supplied gene sets are normally generated by selecting genes that pass a significance threshold. The DAVID procedure is similar to others available such as Ingenuity, AmiGO and GeneGO. The selection of significance values is largely arbitrary, but it is common to set the threshold at FDR adjusted p<0.05. Modifying the threshold will alter the enrichment results considerably. The statistical tests employed include hypergeometric, Fisher's exact and Chi-squared.
In contrast to the above methods that require lists of genes that pass arbitrary thresholds, GSEA is a tool that uses every datapoint in its statistical algorithm. In the "classical" method genes are ranked by from most up-regulated to most down-regulated. The rank metric itself varies, but two valid methods are to use signed p-value, or lower 90% confidence interval of the fold change. The basis of the test is to assess whether members of a gene set appear enriched at one end of the profile. To test the enrichment, GSEA performs permutations of the profile, calculating the enrichment of the gene set a thousand or more times to estimate p-values empirically. Other tools that use the entire profile perform analytical tests like the Wilcoxon and Kolmogorov-Smirnov test.
So now that we understand the main differences in the algorithms, we can start to compare the results from these two techniques. I revisited a dataset I've discussed before, azacitidine treatment in AML3 cells profiled by RNA-seq. I will use the differential RNA-seq expression data generated by DESeq as a start point to compare DAVID and GSEA.
For DAVID, I made a background file consisting of all 19,030 genes detected in the experiment above the detection threshold. The significant (FDR<0.05) gene lists in the up (2502 genes) and down (2907 genes) direction were separately submitted to DAVID 6.7 for analysis, focusing on KEGG pathways only.
For GSEA, I used the same rank metric procedure described previously. I used the classical GSEA algorithm with the gsea2-2.2.2.jar executable, using KEGG gene sets from MSigDB v5.1.
In the graph below, I determined the overlap of statistically significant gene sets (FDR<0.05) separately for up and down regulated pathways. I also included an intersection of DAVID results when no background is provided (for curiosity only, not a recommended procedure).
You'll notice that in both the up and down regulated direction, GSEA identifies more significant results as compared to DAVID. I found this was also the case in other gene set libraries such as REACTOME and GO. The difference in the sensitivity between the two tools boils down to the different algorithms, with DAVID using a discrete list of genes, versus GSEA that uses every data point. GSEA will be much more sensitive in identifying gene sets that have many members that are undergoing subtle changes in expression. Individually, these changes are not statistically significant, but when a large fraction of genes in a set collectively shift in expression, that trend could be very significant statistically speaking.
In addition to the more sensitive algorithm, DAVID's gene sets and gene annotations are really out of date. The last update of DAVID's many data sources being in mid to late 2009. As such, many gene accessions from Ensembl may not be supported. Indeed, from my background file that contained 19,030 only 13,488 were recognized by DAVID. The lack of updated gene sets is also a concern, as is the ever growing number of citations that DAVID still receives.
In conclusion, I would recommend researchers use tools that look at an entire profile to determine enrichment (GSEA, CAMERA, WilcoxGST, Pathifier) rather than submitting gene sets for overlap analysis. Using DAVID might be the quick and easy option, but this comes at the expense of a massive reduction in sensitivity.
Further reading
JA Timmons, KJ Szkop, IJ Gallagher. Multiple sources of bias confound functional enrichment analysis of global-omics data. Genome biology, 2015. 16:186
Lina Wadi, Mona Meyer, Joel Weiser, Lincoln D Stein, Juri Reimand. Impact of knowledge accumulation on pathway enrichment analysis. bioRxiv. doi: http://dx.doi.org/10.1101/049288
EDIT: In May 2016, DAVID was updated!
https://david.ncifcrf.gov/content.jsp?file=release.html
The concepts behind the two algorithms are very different. DAVID determines overlaps between user-supplied gene lists and the curated databases, looking for overlaps that are bigger than that expected by random chance. You can improve the accuracy of the algorithm by providing a background file that contains all genes that were considered/detected in the experiment. The user-supplied gene sets are normally generated by selecting genes that pass a significance threshold. The DAVID procedure is similar to others available such as Ingenuity, AmiGO and GeneGO. The selection of significance values is largely arbitrary, but it is common to set the threshold at FDR adjusted p<0.05. Modifying the threshold will alter the enrichment results considerably. The statistical tests employed include hypergeometric, Fisher's exact and Chi-squared.
In contrast to the above methods that require lists of genes that pass arbitrary thresholds, GSEA is a tool that uses every datapoint in its statistical algorithm. In the "classical" method genes are ranked by from most up-regulated to most down-regulated. The rank metric itself varies, but two valid methods are to use signed p-value, or lower 90% confidence interval of the fold change. The basis of the test is to assess whether members of a gene set appear enriched at one end of the profile. To test the enrichment, GSEA performs permutations of the profile, calculating the enrichment of the gene set a thousand or more times to estimate p-values empirically. Other tools that use the entire profile perform analytical tests like the Wilcoxon and Kolmogorov-Smirnov test.
So now that we understand the main differences in the algorithms, we can start to compare the results from these two techniques. I revisited a dataset I've discussed before, azacitidine treatment in AML3 cells profiled by RNA-seq. I will use the differential RNA-seq expression data generated by DESeq as a start point to compare DAVID and GSEA.
For DAVID, I made a background file consisting of all 19,030 genes detected in the experiment above the detection threshold. The significant (FDR<0.05) gene lists in the up (2502 genes) and down (2907 genes) direction were separately submitted to DAVID 6.7 for analysis, focusing on KEGG pathways only.
For GSEA, I used the same rank metric procedure described previously. I used the classical GSEA algorithm with the gsea2-2.2.2.jar executable, using KEGG gene sets from MSigDB v5.1.
In the graph below, I determined the overlap of statistically significant gene sets (FDR<0.05) separately for up and down regulated pathways. I also included an intersection of DAVID results when no background is provided (for curiosity only, not a recommended procedure).
 |
| DAVID vs GSEA using the same gene expression profile. Numbers represent gene sets that are significantly enriched (FDR<0.05). |
In addition to the more sensitive algorithm, DAVID's gene sets and gene annotations are really out of date. The last update of DAVID's many data sources being in mid to late 2009. As such, many gene accessions from Ensembl may not be supported. Indeed, from my background file that contained 19,030 only 13,488 were recognized by DAVID. The lack of updated gene sets is also a concern, as is the ever growing number of citations that DAVID still receives.
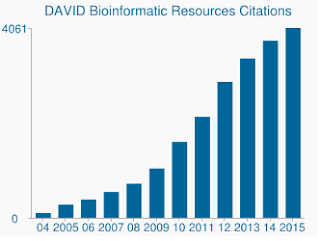 |
| DAVID still receives thousands of citations per year despite underlying data being woefully out of date. |
Further reading
JA Timmons, KJ Szkop, IJ Gallagher. Multiple sources of bias confound functional enrichment analysis of global-omics data. Genome biology, 2015. 16:186
Lina Wadi, Mona Meyer, Joel Weiser, Lincoln D Stein, Juri Reimand. Impact of knowledge accumulation on pathway enrichment analysis. bioRxiv. doi: http://dx.doi.org/10.1101/049288
EDIT: In May 2016, DAVID was updated!
https://david.ncifcrf.gov/content.jsp?file=release.html

