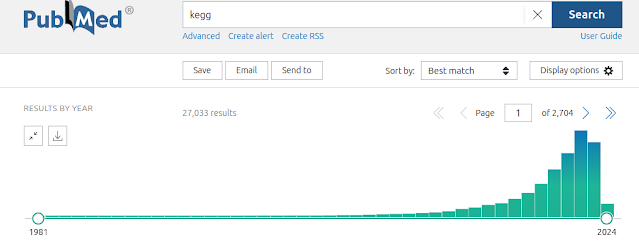
KEGG is the Kyoto Encyclopedia of Genes and Genomes, a compendium of gene functional annotations which are commonly used for pathway enrichment analysis. KEGG has been around since 1997 (PMID: 9390290) but in 2000, as the draft human genome sequence was released, KEGG became a key database involved in the curation of literature data to catalog the function of all human genes, many of which were newly sequenced and previously uncharacterised. This invigorated interest in KEGG is demonstrated by one of their articles in 2000 (PMID: 10592173) accruing 24,236 citations according to Dimensions, 1270 fold higher than other articles from that time. A PubMed search using "KEGG" keyword shows 27,033 results, with matches in abstracts alone and the tragectory is increasing at a rapid rate. Despite the popularity of this tool, I'm urging you not to use it for your research. Here I will lay out the reasons.
1. It isn't comprehensive
I did an analysis of KEGG and other pathway sets in
MSigDB and found some interesting results. Not only does KEGG have fewer gene sets, but the number of unique genes described is very small compared to Reactome or Gene Ontology Biological process. This is likely to bias over-representation based enrichment tests as genes without annotations are routinely excluded from an analysis by some tools. The total number of annotations was seven fold bigger for Reactome compared to KEGG legacy and 49 fold for GO BP. Sure, Reactome and GO BP have a lot of redundancy caused by the hirarchical structure and some sets with different names and similar member genes, but it is clear from this data that KEGG really isn't comprehensive as compared to the alternatives. If you want to see how this was done, you can download my script
here.
2. Restrictive licence and cost
KEGG may be free to use in some situations, but it is clear that for non-academic users, it is a pay-to-play proposition (
link). Moreover academic users may not directly access the FTP data store, presumably to prevent unauthorised data mining. Even academic service providers need to obtain a subscription, which costs USD$5,000 per year if accessed by more than one individual. Moreover, it is unclear from the licence whether subscribers are able to disseminate large scale data derived from the KEGG database. Lastly, if the subscription is terminated, then users must legally delete any derivative data including ML/AI models (
link). In contrast Gene Ontology provides their data under a relatively permissive Creative Commons Attribution 4.0 Unported License, which provides a high degree of flexibility for users to access, analyse and remix GO data. Reactome has a CC0 licence which essentially puts its data into the public domain.
3. Lack of archival versions
Ms Anusuiya Bora, a PhD candidate in my lab reached out to the KEGG administrators to request some historical KEGG gene sets which were a part of a tool called DAVID version 6.8. Sadly DAVID 6.8 has been retired by the administrators, and so roughly 20,000 articles in PubMed are no longer reproducible with the original tools. A lot of these articles were published from 2018-2021, and so it is disappointing that we can't go back and check them. Even the authors themselves won't be able to reproduce their work, despite the fact that many institutions and funders require researchers retain their data and algorithms for 5 years or more after publication. On the other hand, Gene Ontology has archival data dumps that go back to 2004 (
here). Reactome has earlier versions accessible by MySQL queries (
link).
Conclusion
KEGG enjoys a big user base due to being an early mover into the pathway annotation space, but the coverage has fallen behind other initiatives like Gene Ontology and Reactome which have large-scale support from the NHGRI and EMBL. KEGG appears to have pivoted towards a commericial direction to raise funds to maintain the curation work, but it is struggling to compete. The lack of archival KEGG versions is a major blunder and reminiscent of the
loss of the original Apollo 11 video recordings. Indeed, it would be nice to look back on the early growth of pathway knowledge in the early 2000's, but this won't be possible for KEGG. This highlights that longer term reproducibility is going to be easier when the tools and data are free and open source. They can be archived permanently in Zenodo or Software Heritage, and future reproducers don't need to agree to unreasonable conditions or fees for access. Finally, when such resources are controlled by a commercial company, it cannot be guaranteed that they will continue to operate, if they go bust, these resources might be gone forever.
I would like to thank Ms Anusuiya Bora who collected a lot of the information shown here.
Comments and discussion on Mastodon here: https://genomic.social/@mdziemann/111933336579691473