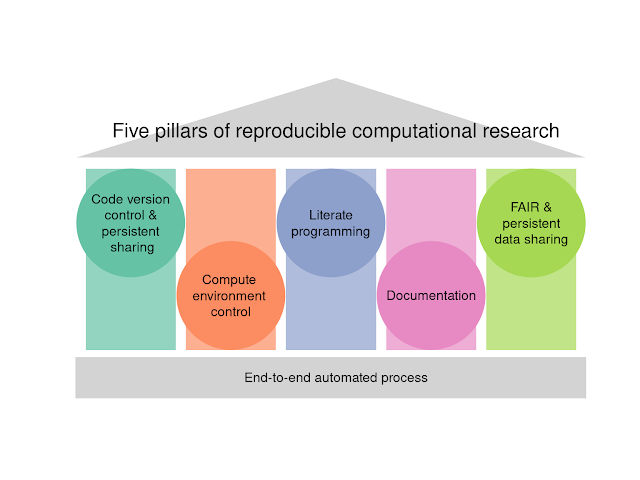
The five pillars of computational reproducibility: Bioinformatics and beyond
I've been working on a new project to follow-up our paper last year on the problems with pathway enrichment analysis. That article turned out to be a bleak and depressing look into how frequently used tools in genomics are misused. It is not an exaggeration to say that most articles showing some type of enrichment analysis are doing it wrong and no doubt this is severely impacting the literature.
However I think it isn't helpful to only focus on the negative aspects of bioinformatics and computational research. We also need to lead the way towards resolving these issues. The best way to do this is in my view is to provide step-by-step guides and tutorials for common routines. So this is what we are in the process of doing, making a protocol for pathway enrichment analysis that is "extremely reproducible". By this, I mean that the analysis could be reproduced independently in future with the minimum of fuss and time. As we were writing this we also recognised that the overall philosophy we devised had not really been generalised yet, and would probably be of interest for researchers doing things outside of genomics.
We drafted the infographic with the five pillars and sought feedback and contributions from our contacts. Then with the coauthors Pierre Poulain (Université Paris Cité) and Anusuiya Bora (my student at Deakin Uni) we elaborated the infographic into a review that gives the full picture of current best practices in computational research. The preprint is online at OSF here.
In the spirit of the content of the review, the document itself is a reproducible R Markdown document, able to be executed using a R docker image, with the code living on github version control here along with helpful documentation courtesy of a detailed README.If you have feedback on the preprint, feel free to raise an issue on GitHub. It is appreciated.
Thanks to Dr Aziz Khan (Stanford University), Dr Sriganesh Srihari (QIMR Berghofer Medical Research Institute), Dr Barbara Zdrazil (EMBL-EBI) and Dr Irene López Santiago (EMBL-EBI) for comment on the five pillars idea. Special thanks to Dr Altuna Akalin (Max Delbrück Center, Germany), Dr Simon Tournier (Paris Cité University, France), Dr Samuel S. Minot (Fred Hutchinson Cancer Center) and Dr Martin O’Hely (Deakin University) for feedback, comments and advice.