DEE2 project update October 2020
Bit of a thread for some updates to the DEE2 data set. It's a resource of uniformly processed RNA-seq data free to use under a permissive GPL3 licence. Find it at http://dee2.io
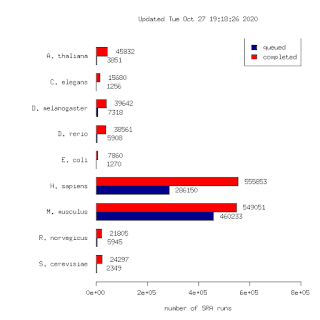
Yesterday a batch of 117k human runs were uploaded. This brings the total number of runs to 1,298,581. To my knowledge this is the largest such data set in the world. This is 10x larger than our first release in 2015! (125k runs)
The number of SRA projects with completed data analysis bundles is 32692
Accesible here: http://dee2.io/huge/
The getDEE2 package is the recommended way to access this data if you are familiar with R. You can access individual runs or enrire data bundles with the various functions. The pkg is part of the latest BioC release out today https://www.bioconductor.org/packages/devel/bioc/html/getDEE2.html
The button for redirecting DEE2 data directly to Degust is broken and we are looking for a fix. For now you will need to download the data to the disk and upload to the Degust webpage: https://degust.erc.monash.edu/
Search capability has always been a bit underwhelming with the DEE2 web interface so we are working on a more modern approach to this in the next couple of months.
We always hoped that DEE2 data could be used to generate canonical gene signatures. We have completed a pilot study with budding three undergrad biocurators who focused on generating gene sets related to our research focus: cardiovascular disease, diabetes, epilepsy and COVID-19. http://dee2.io/genesets.html
The time between SRA deposition and DEE2 has been something like 3-6 months. Too long! We will be focusing on bringing this down to about 1 month. If we receive an additional HPC grant we will be able to shrink this to a few weeks. If that grant is funded we will also be in a good place to expand DEE2 to encompass a number of other species including some crops like wheat, maize and tomato, as well as animals such as cattle, chicken and macaques.
Further ahead we are aware that the new mouse genome assembly GRCm39 will be annotated and part of the Ensembl release soon so we will be looking for additional compute to reproces all the mouse data on the new assembly in the next 12 months.
Although the dataset is GPL3, if you use the dataset for financial gain, you should consider sponsoring the project. It will help the long term viability, expansion and new features and improvements.
I'd like to thank a few people:
- Antony Kaspi at WEHI for his work on the project
- Undergrad biocurators Aaron Kovacs, Chelsia Sritharan and Haris Lekovic
- Computational partners at MASSIVE, Deakin eResearch and Nectar/ARDC
- Users for your suggestions and encouragement