Update on DEE2 project for Sept 2018
A few updates for DEE2 i would like to share.
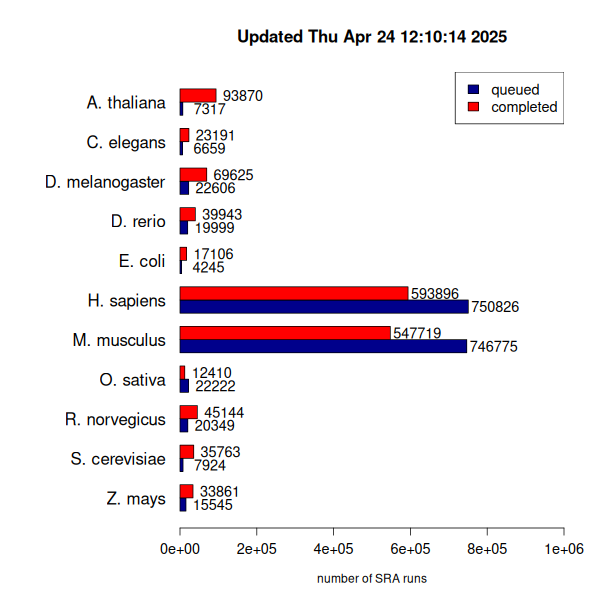
- I switched over to NameCheap domain name service which appears to be working much nicer than the previous one (HostPapa). The domain name sever change broke the docker image so it was slightly modified and rebuilt.
- I've integrated with SRAdbV2, an now there are many more datasets in the queue. I think many of these are small ones related to single cell RNA-seq. I am using as many computers as possible to clear up the backlog. I've noticed a lot of SRA project with one or a few datasets missing, so I have have written a script to identify these and queue them with priority.
- The R interface hs undergone several improvements and should be more robust now. A whole bunch of new documentation has been added, including a complete walkthrough starting with SRAdbV2 query, fetching DEE2 data, and differential analysis with edgeR and DESeq.
- Also bulk data dumps are again available via http. Dat turned out to be too slow and unreliable for files of this size unfortunately.
The priorities going forward are:
- Improve QC info:
- Link to QC info in the search results. Currently users need to download the expression data to browse the quality metrics.
- Currently all datasets are "PASS" but I will need to classify "WARNING" and "FAIL" as well.
- Transfer live data to Deakin hosted infrastructure.
- Finish the data processing and manscript.

