Introducing the ENCODE Gene Set Hub
TL;DR We curated a bunch of ENCODE data into gene sets that is super useful in pathway analysis (ie GSEA).
Link to gene sets and data: https://sourceforge.net/projects/encodegenesethub/
Poster presentation: DOI:10.13140/RG.2.2.34302.59208
Now for the longer version. Gene sets are wonderful resources. We use them to do pathway level analyses and identify trends in data that lead us to improved interpretation and new hypotheses. Most pathway analysis tools like GSEA allow us to use custom gene sets, this is really cool as you can start to generate gene sets based on your own profiling work and that of others.
There is huge value in curating experimental data into gene sets, as the MSigDB team have demonstrated. But overall, these data are under-shared. Even our group is guilty of not sharing the gene sets we've used in papers. There have been a few papers where we've used gene sets curated from ENCODE transcription factor binding site (TFBS) data to understand which TFs were driving responses to various stimuli [1-5]. Coauthors and I realised the need for our ENCODE gene set resource to be public. Our goal is to release these gene sets (and code) as a public resource and write a paper demonstrating the utility of this resource in enhancing interpretation of experimental data.
In this first piece of work, we selected ENCODE data that are present online in "peak" format, which includes TFBS, histone mods and DNase-seq peaks. We have curated lists of genes that contain such peaks at transcription start sites as well as enhancers.
The end goal, to publish really high quality gene sets, pushed us to think about ways to optimise gene set curation and how to even measure gene set quality. There was not a lot of literature to base our work off. We reasoned that real gene expression profiles are strongly non-random as we know clusters of co-regulated genes exist. The average enrichment score (ie NES for GSEA) of all genes in a set could be useful in detecting non-random sets of genes. The problem lies in that as you change parameters, the number of members of the gene sets change, and as you add more members to a set, the NESs tend to increase and raw enrichment scores (ESs) tend to decrease. So we reasoned that we should somehow normalise for the number of genes in the set. We devised a metric that was useful in estimating association between gene sets and profiling data:
AM=(NES^2) / Size
Where AM is the association metric, NES is the normalised enrichment score for each gene set and "Size" is the number of genes in the gene set. In our work, the average AM of all sets in the GSEA run is reported. The higher the AM, the less random the gene sets are. In the process of turning peak data into gene sets we came into a few key questions:
Using this approach as shown in our poster presented at the 2017 Lorne Genome Conference we were able to define AM while varying parameters like the size of the gene set and distance of peaks to the regulatory landmarks.
We found that gene sets of 2000 or more should be avoided, and the distance between peak and regulatory landmark can be 1 kbp to 10 kbp. Results were consistent for TFBS, histone mod and DNase-seq peaks at both TSSs and distal regulatory elements.
While there are already curated TF binding site data from ENCODE at Harmonizome, these are not readily accessible in bulk. Our work is unique for the following reasons
MultiRes can be used for other layers of regulation, such as Starbase microRNA target gene sets.
In the next iteration of the Encode Gene Set Hub, we will be adding gene sets from FAIRE-seq, RNA-seq and other ENCODE data.
If you use these gene sets in your work, I'd love to hear some feedback. leave your comments below.
References
Link to gene sets and data: https://sourceforge.net/projects/encodegenesethub/
Poster presentation: DOI:10.13140/RG.2.2.34302.59208
Now for the longer version. Gene sets are wonderful resources. We use them to do pathway level analyses and identify trends in data that lead us to improved interpretation and new hypotheses. Most pathway analysis tools like GSEA allow us to use custom gene sets, this is really cool as you can start to generate gene sets based on your own profiling work and that of others.
There is huge value in curating experimental data into gene sets, as the MSigDB team have demonstrated. But overall, these data are under-shared. Even our group is guilty of not sharing the gene sets we've used in papers. There have been a few papers where we've used gene sets curated from ENCODE transcription factor binding site (TFBS) data to understand which TFs were driving responses to various stimuli [1-5]. Coauthors and I realised the need for our ENCODE gene set resource to be public. Our goal is to release these gene sets (and code) as a public resource and write a paper demonstrating the utility of this resource in enhancing interpretation of experimental data.
In this first piece of work, we selected ENCODE data that are present online in "peak" format, which includes TFBS, histone mods and DNase-seq peaks. We have curated lists of genes that contain such peaks at transcription start sites as well as enhancers.
The end goal, to publish really high quality gene sets, pushed us to think about ways to optimise gene set curation and how to even measure gene set quality. There was not a lot of literature to base our work off. We reasoned that real gene expression profiles are strongly non-random as we know clusters of co-regulated genes exist. The average enrichment score (ie NES for GSEA) of all genes in a set could be useful in detecting non-random sets of genes. The problem lies in that as you change parameters, the number of members of the gene sets change, and as you add more members to a set, the NESs tend to increase and raw enrichment scores (ESs) tend to decrease. So we reasoned that we should somehow normalise for the number of genes in the set. We devised a metric that was useful in estimating association between gene sets and profiling data:
AM=(NES^2) / Size
Where AM is the association metric, NES is the normalised enrichment score for each gene set and "Size" is the number of genes in the gene set. In our work, the average AM of all sets in the GSEA run is reported. The higher the AM, the less random the gene sets are. In the process of turning peak data into gene sets we came into a few key questions:
- Does it matter how many genes there are in a set?
- Does it matter how far away a peak is from a TSS or enhancer (regulation landmark)?
- Which gene annotation set we should use?
Using this approach as shown in our poster presented at the 2017 Lorne Genome Conference we were able to define AM while varying parameters like the size of the gene set and distance of peaks to the regulatory landmarks.
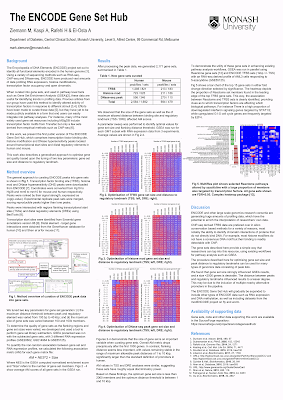 |
| Our poster for Lorne Genome Conference 2017 (DOI:10.13140/RG.2.2.34302.59208) |
While there are already curated TF binding site data from ENCODE at Harmonizome, these are not readily accessible in bulk. Our work is unique for the following reasons
- Our analysis looks at TFBS at enhancers as well as promoters
- Our work summarises mouse ENCODE data
- Gene set size and peak-landmark distance are optimised
- Includes chromatin accessibility and histone mod data in addition to TFBS
During the course of this work, we also developed MultiRes which is a visualisation tool for multiple layers of regulation using ComplexHeatmap/R. For instance, in the poster, we used Reactome pathways as a primary analysis (red and green) and layered ENCODE transcription factor binding site data over as a secondary analysis (blue, white and red).
 |
| MultiRes plot showing Reactome and ENCODE TFBS analyses. |
In the next iteration of the Encode Gene Set Hub, we will be adding gene sets from FAIRE-seq, RNA-seq and other ENCODE data.
If you use these gene sets in your work, I'd love to hear some feedback. leave your comments below.
References
