microRNA aligners compared
Alignment of microRNA to the genome poses a particular challenge because the reads are short, and some microRNAs are nearly identical. Moreover, microRNAs themselves are subject to RNA editing (adenine-to-inosine conversion, non-templated base addition) and normal sequencing error rates. In this post, I'm going to test the performance of several aligners in aligning microRNA reads to the Arabidopsis genome.
I downloaded the Arabidopsis genome from Ensembl plant and the latest miRbase release version 21. I used bowtie2 to align the 325 full-length hairpin transcripts to the Arabidopsis genome. I generated pseudo microRNA reads that uniformly cover the hairpin transcript at a range of lengths from 16 nt to 25 nt. I then aligned the reads to the Arabidopsis genome using these different aligners with the default settings. I then used bedtools and awk to count the correctly and incorrectly mapped reads at a mapQ threshold of 10.
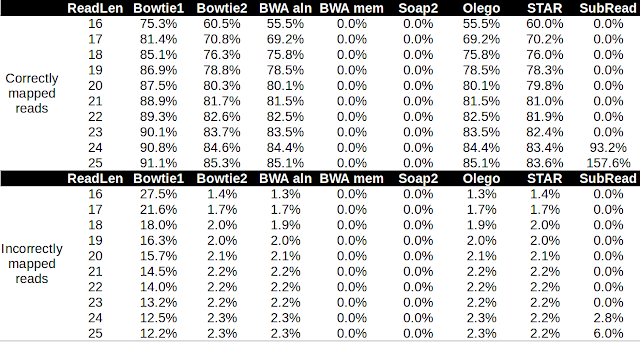 |
| Table 1. Performance of several aligners in pseudo microRNA read alignment to the Arabidopsis genome. |
Firstly note that there were no alignment with either Soap2 or BWA mem and so these aligners are not suitable for microRNA with the default settings. SubRead only started generating alignments at 24 nt in length so again, not suitable for microRNA with the default parameters. I graphed the performance of Bowtie1, Bowtie2, BWA aln, OLego and STAR.
|
Figure 1 shows that Bowtie1 has the highest correct mapping rate, but this comes at a cost of increased incorrect mapping. Bowtie2, STAR, BWA aln and OLego showed fairly similar results with 21mers aligned correctly ~81% and incorrectly ~2.2% of the time.
To simulate the challenges with real-world situations where the reads may have sequencing errors, RNA editing and bona fide SNVs, I mutated the reads using EMBOSS msbar. I incorportated up to 3 SNPs, and up to 2 insertions or deletions.
.png) | |
| Figure 2. Mutated pseudo microRNA read alignment to the Arabidopsis genome. |
Next, I incorporated up to 2 random nucleotides to the 3 prime or 5 prime ends and performed the same test.
 |
| Figure 3. End-modified pseudo microRNA read alignment to the Arabidopsis genome. |
Then I summarised the results by calculating the average % correct/incorrect mapping rates for the different tests in the range of read lengths (16 to 25 nt).
 |
| Table 2. Aligner performance for mutated pseudo microRNA reads to the Arabidopsis genome. Read length analysed was 16 to 25 nt. |
These test show that Bowtie1 is quite inaccurate, even for exact-matching reads and so I would caution against using it for microRNA. Of the remaining four aligners in this test, Bowtie2 showed the lowest incorrect mapping rates at the cost of lower correct mapping of mutated reads. BWA aln, OLego and STAR had very similar performance.
My follow up to this work was recently published in RNA Journal!
My follow up to this work was recently published in RNA Journal!


