Update your gene names when doing pathway analysis of array data!
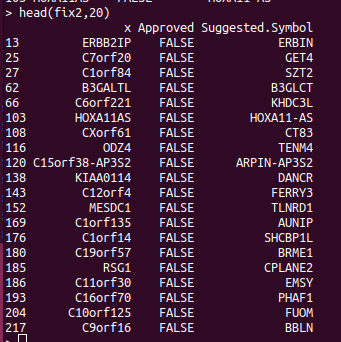
If you are doing analysis of microarray data such as Infinium methylation arrays, then those genomic annotations you're using might be several years old. The EPIC methylation chip was released in 2016 and the R bioconductor annotation set hasn't been updated much since. So we expect that some gene names have changed, which will reduce the performance of the downstream pathway analysis. The gene symbols you're using can be updated using the HGNChelper R package on CRAN. Let's say we want to make a table that maps probe IDs to gene names, the following code can be used. library("IlluminaHumanMethylationEPICanno.ilm10b4.hg19") anno <- getAnnotation(IlluminaHumanMethylationEPICanno.ilm10b4.hg19) myann <- data.frame(anno[,c("UCSC_RefGene_Name","UCSC_RefGene_Group","Islands_Name","Relation_to_Island")]) gp <- myann[,"UCSC_RefGene_Name",drop=FALSE] gp2 <- strsplit(gp$UCSC_RefGene_Name,";") names...